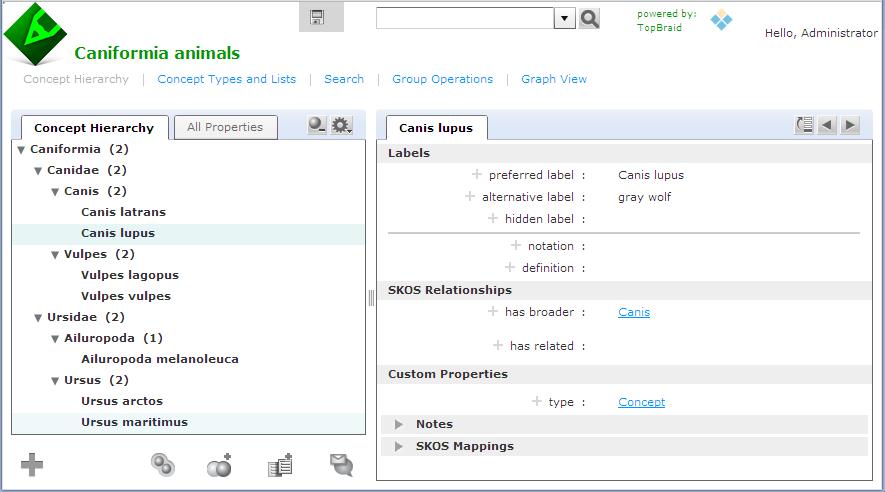
In an earlier entry, we learned how SPARQL Rules can increase the quality of taxonomies and other controlled vocabularies stored using the W3C SKOS ontology. (As I wrote there, the Simple Knowledge Organization Systemvocabulary management specification is gaining popularity because, as a standard, it makes it easier to share taxonomies and thesaurii between different systems. It also guards investments in vocabulary development against the potential problems of dependence on a proprietary vendor format.) TopQuadrant's Enterprise Vocabulary Net (EVN) vocabulary manager uses SKOS as its default format for storing data. Whether you use EVN or not, a first step in systematic management of vocabularies is often the conversion of vocabularies stored in ad hoc spreadsheets—an unfortunately very popular way to store them—to SKOS, so today we'll look at how TopBraid makes this conversion easy. Below is an Excel spreadsheet with some data about a few Caniformia animals. (In the Linnaeus classification of animals, Caniformia is the suborder of Carnivora, which is an order of the Mammalia class.) It shows two families of this suborder and a few genuses and species of each family, with both the Latin and common name of each species. Using a SPARQLMotion script, the basic steps of converting a spreadsheet like this to SKOS are: Read in the spreadsheet as a set of RDF triples. Use a CONSTRUCT query to convert the spreadsheet triples to SKOS triples. This is the step that varies the most from one conversion to another, because people can arrange spreadsheets any way they want, so the logic of the CONSTRUCT query has to infer the correct relationships between the values on the spreadsheet. Save the SKOS triples as an RDF file or in whatever format is appropriate to your applications that will use this data. The following shows the SPARQLMotion script that I used to convert the spreadsheet above. It has a module for each of the three steps listed above and an additional SetBaseURIStr module to set a ?baseURIStr variable. The script refers to the base URI of the output several times, and instead of hardcoding it in all those places, I decided to use this module to set this variable and to then reference the variable from other places so that resetting the base URI could be done in one place. The "set BaseURIString" module has a very simple SELECT query: When you import an Excel file into TopBraid, the "Import Excel Cell Instances" SPARQLMotion module can pull triples from the spreadsheet with information such as the fact that a given cell has a row value of 7 (using zero-based counting), a column value of 0, a type value of "xsd:string", and "giant panda" as its contents. This level of detail can be useful for picking apart complex spreadsheets, but for simpler ones, if you instead use an "Import RDF from Workspace" module (in other words, if you have the script open the spreadsheet as if it were an RDF file), TopBraid uses the headings of the spreadsheet to identify more of the semantics of the data. For example, it would create triples saying that the thing identified as Row-6 has a commonName value of "giant panda" and a genus value of "Ailuropoda". This will be easier to convert to SKOS with a CONSTRUCT query. There are five basic tasks that the conversion module must perform, all through the creation of triples: Declare that the dataset being created is an ontology. Import the standard W3C SKOS ontology so that we can reference its classes and properties. Declare a concept scheme. A SKOS vocabulary can have as many concept schemes as you like, but we'll just create one for our example. Declare concepts for each species, genus, and family found in the input triples, with a skos:broader property pointing from each one to either the appropriate broader concept or, if there is none, to the concept scheme created in the previous step. Create triples that attach any additional metadata to the appropriate concepts—in this case, to assign the common name value to each species concept. SKOS is very flexible, so if you had additional non-SKOS properties specific to your own applications that you wanted to assign to each concept, the steps would be similar to the ones for attaching the common name values from this spreadsheet to each concept. Whe the "Import RDF from Workspace" module reads in a spreadsheet such as caniformia.xls, it uses the spreadsheet's filename to define a prefix for the spreadsheet's properties so that it can refer to those properties with names like caniformia:genus. After opening the spreadsheet directly in TopBraid Composer, I saw that the base URI created for the data and associated with the caniformia: prefix was file:///xls2skos/caniformia.xls, because I had it in a project named xls2skos. I wanted to use this prefix in my SPARQLMotion script's CONSTRUCT query, so I associated this URI with the caniformia: prefix in the Overview tab of the xls2skos.n3 file that stored the SPARQLMotion script. The actual conversion takes place in the Apply Construct module that I named "convert XLSData". These modules can store multiple CONSTRUCT queries, so I used two. The first does the basic setup of the taxonomy being created, which are the first three of the five tasks listed above: The second query performs steps 4 and 5: In addition to creating a SKOS concept for each species, it creates one for each genus and family as well, using the skos:broader property to identify the connections between these concepts that make up the hierarchical taxonomy of terms. One big decision to make with this query was how to create URIs that provided unique identifiers for each new concept being created. I knew that the species, family, and genus names must be unique, so I added those to the base URI after passing them to smf:encodeURL(), a SPARQLMotion extension function that escapes any characters that won't work well in a URI. If you have taxonomy data in a spreadsheet, there may already be a unique number or other form of ID assigned to some or all taxonomy terms that your conversion can grab so that you don't have to create URIs from the names on the spreadsheet like I did. I also decided to use species names like "Canis lupus" as the skos:preferredLabel value in the output and to use labels from the spreadsheet's "common name" column like "gray wolf" as skos:altLabel values. If you wanted to to use common name values as preferred labels and species names as alternative labels, it would be a simple change to the query above. My "convert XLSData" module also has its sml:replace value set to True so that it doesn't pass along the input triples to the final module, which saves the conversion result. This last module, which I named "Save as TDB", is an "Export to TDB" SPARQLMotion module that saves the conversion results using the Jena TDB format. I could have used a SPARQLMotion "Export to RDF" module, which saves triples as a Turtle or RDF/XML disk file, but I wanted to use the results of my conversion in EVN. EVN requires that you use Jena's TDB or SDB formats so that it can attach metadata to your work to support reporting and workflow tracking. After running the conversion, here is one view of it in EVN: This is a minimal example using a small spreadsheet with no non-SKOS metadata. If your spreadsheet includes columns for data that don't fit easily into the SKOS model, you can use TopBraid Composer to create a customized version of SKOS that includes your own properties, and your SPARQLMotion script's conversion module can then add triples for those properties to the result. Viewing them in EVN, they would appear under Custom Properties on the right. And of course, the screen shot above only hints at all that EVN lets you do with your controlled vocabulary once you convert it to SKOS.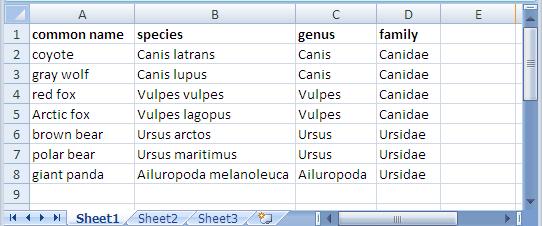

SELECT ?baseURIStr
WHERE {
LET (?baseURIStr := "http://example.com/taxonomies/animals") .
}CONSTRUCT {
?baseURI a owl:Ontology .
?baseURI owl:imports <http://www.w3.org/2004/02/skos/core> .
<http://example.com/taxonomies/animals/caniformia> a skos:ConceptScheme .
<http://example.com/taxonomies/animals/caniformia> rdfs:label "Caniformia" .
}
WHERE {
LET (?baseURI := smf:buildURI("<{?baseURIStr}>")) .
}CONSTRUCT {
?speciesURI a skos:Concept .
?genusURI a skos:Concept .
?familyURI a skos:Concept .
?speciesURI skos:prefLabel ?speciesName .
?speciesURI skos:altLabel ?commonName .
?speciesURI skos:broader ?genusURI .
?genusURI skos:broader ?familyURI .
<http://example.com/taxonomies/animals/caniformia> skos:hasTopConcept ?familyURI .
}
WHERE {
?row caniformia:commonName ?commonName .
?row caniformia:species ?speciesName .
LET (?species := smf:encodeURL(?speciesName)) .
LET (?speciesURI := smf:buildURI("<{?baseURIStr}#{?species}>")) .
?row caniformia:genus ?genusName .
LET (?genus := smf:encodeURL(?genusName)) .
LET (?genusURI := smf:buildURI("<{?baseURIStr}#{?genus}>")) .
?row caniformia:family ?familyName .
LET (?family := smf:encodeURL(?familyName)) .
LET (?familyURI := smf:buildURI("<{?baseURIStr}#{?family}>")) .
}
Wednesday, December 29, 2010
How to: convert a spreadsheet to SKOS
Posted by
Bob DuCharme
at
11:10 AM
0
comments
![]()
Subscribe to:
Posts (Atom)
This is a blog by TopQuadrant, developers of the TopBraid Suite, created to support the pursuit of our ongoing mission - to explode strange semantic myths, to seek out new models that support a new generation of dynamic business applications, to boldly integrate data that no one has integrated before.




